Transcription Regulation in Pathogenic Bacteria
Approximately one-third of the world population is infected with Mycobacterium tuberculosis and millions die from this disease annually. The success of this pathogen lies in its ability to lay dormant for long periods of time and to resist the attempts of the immune system to eradicate it from the host. Although much is known about transcription initiation in model bacterial such as Escherichia coli, relatively little is known about the mechanisms of basal transcription in mycobacteria. For example, transcription factors that are essential in M. tuberculosis such as CarD and RbpA are not even present in the genome of E. coli. We have been studying the energetics of transcription initiation in this important pathogen to better understand the diversity of gene expression pathways in bacteria. In addition, as multi-drug resistant strains are becoming more prevalent we hope to find novel molecular strategies to interfere with the unique regulatory pathways of M. tuberculosis and treat this costly disease.
 CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase.
CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase.Jensen D, Ruiz Manzano A, Rammohan J, Stallings CL, Galburt EA
Nucleic Acids Research. 2019 July; 47(13): 6685–6698.
The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression.
Galburt EA
Proceedings of the National Academy of Sciences. 2018 December; 115(50):E11604-E11613.
Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD.
Rammohan J, Ruiz Manzano A, Garner AL, Stallings CL, Galburt EA
Nucleic Acids Research. 2015 February; 43(6):3272-85.
DNA Repair in Mycobacterium tuberculosis
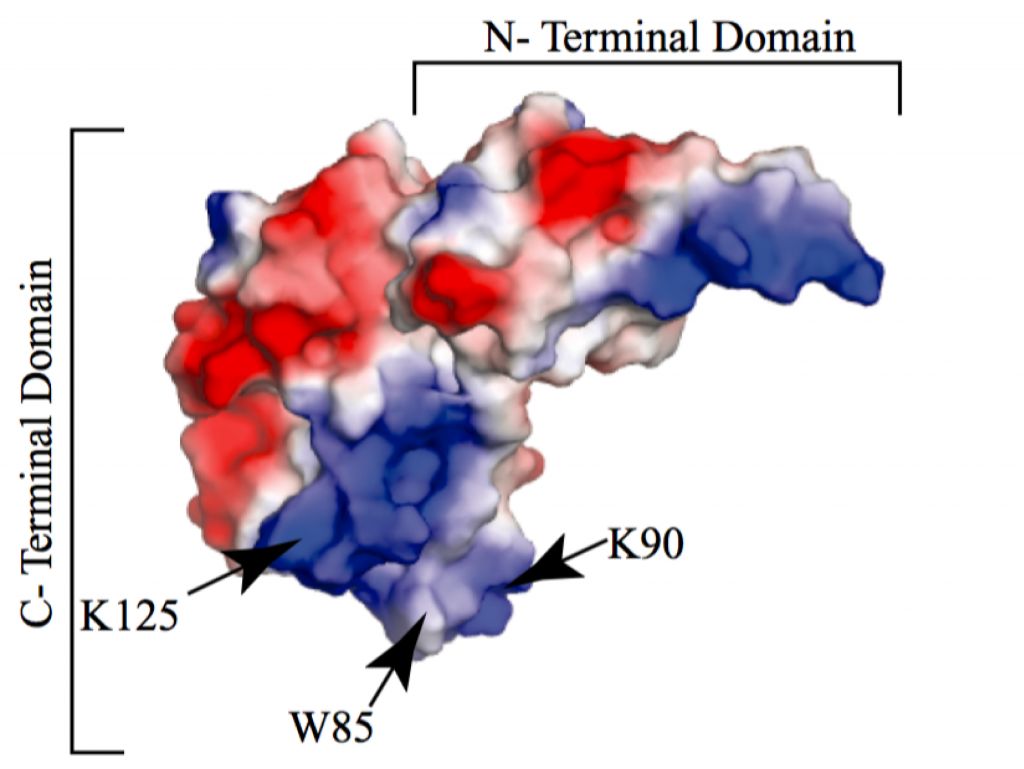
Our studies of the molecular pathways of Mtb have recently expanded to DNA repair. Specifically, we discovered that the activity of a critical DNA repair helicase, UvrD1, is regulated by redox potential. Whereas a monomer of UvrD1 can translocate on single-stranded DNA, it is unable to unwind duplex. In contrast, a disulfide bonded dimer is able to unwind multiple turns of DNA in about a second. Our future work is aimed at understanding how this regulation is enacted in bacterial cells under stress and what are the structural determinants of activation.
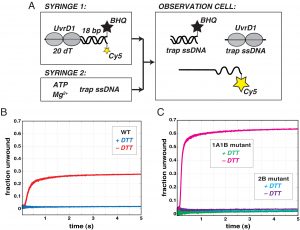
Mycobacterium tuberculosis DNA repair helicase UvrD1 is activated by redox-dependent dimerization via a 2B domain cysteine.
Chadda A, Jensen D, Ruiz Manzano A, Nguyen B, Lohman T, Galburt EA
PNAS. 2022 February; 119(8)
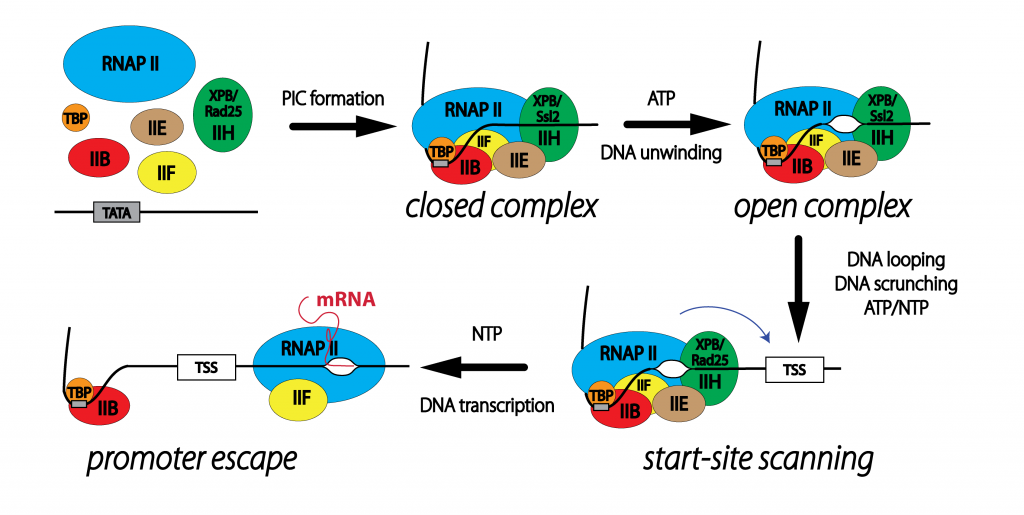
mechanisms of transcription initiation in yeast and humans
The ability of cells to interact and respond to their environment is largely dependent on the regulation of gene expression. Much of this regulation occurs at the stage of transcription initiation where an RNA polymerase is recruited to a promoter, the DNA is unwound, and the polymerase begins processive transcription elongation. In Eukaryotes, basal and activated initiation is dependent on the formation of a "pre-initiation complex" or PIC that is made up of over 30 polypeptide chains and contains a variety of DNA binding and catalytic activities. For example, as many as three different ATPase centers collaborate to produce an elongating polymerase. We are interested in dissecting the molecular mechanism of the PIC to set the stage for a deeper understanding of how this process is regulated during changes in gene expression as well as how the process may go awry in certain disease states.